Ytterbium »
PDB 1c5k-3fty »
1z1y »
Ytterbium in PDB 1z1y: Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
Protein crystallography data
The structure of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax, PDB code: 1z1y
was solved by
A.K.Saxena,
K.Singh,
H.P.Su,
M.M.Klein,
A.W.Stowers,
A.J.Saul,
C.A.Long,
D.N.Garboczi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.60 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.164, 43.700, 65.650, 90.00, 103.15, 90.00 |
| R / Rfree (%) | 24.4 / 27.6 |
Ytterbium Binding Sites:
The binding sites of Ytterbium atom in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
(pdb code 1z1y). This binding sites where shown within
5.0 Angstroms radius around Ytterbium atom.
In total 9 binding sites of Ytterbium where determined in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax, PDB code: 1z1y:
Jump to Ytterbium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Ytterbium where determined in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax, PDB code: 1z1y:
Jump to Ytterbium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
Ytterbium binding site 1 out of 9 in 1z1y
Go back to
Ytterbium binding site 1 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
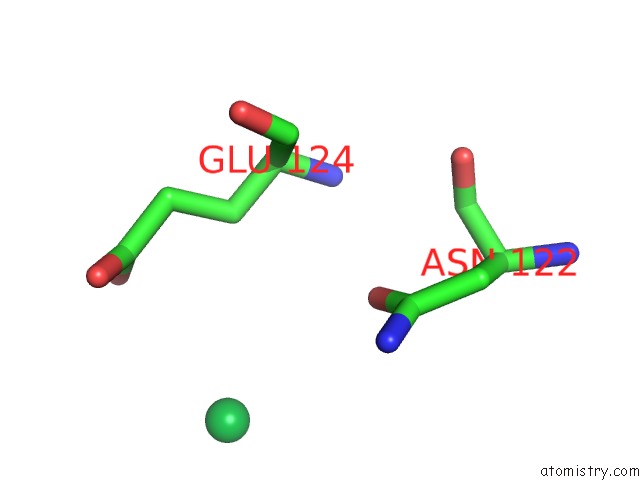
Mono view
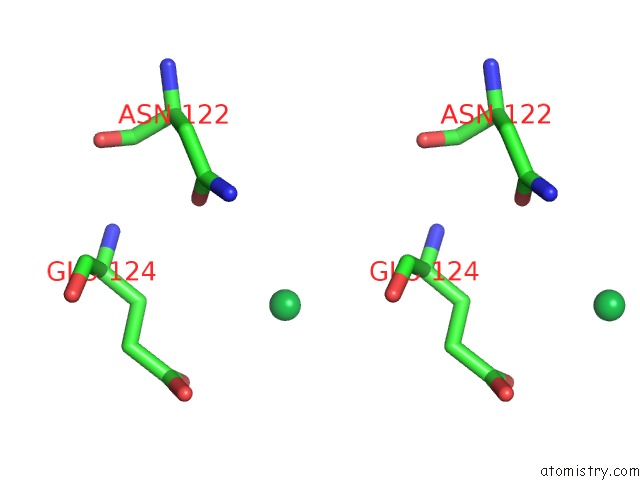
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 1 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 2 out of 9 in 1z1y
Go back to
Ytterbium binding site 2 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
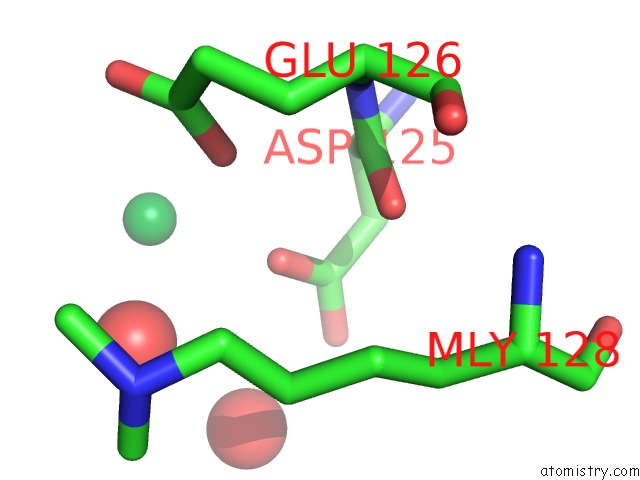
Mono view
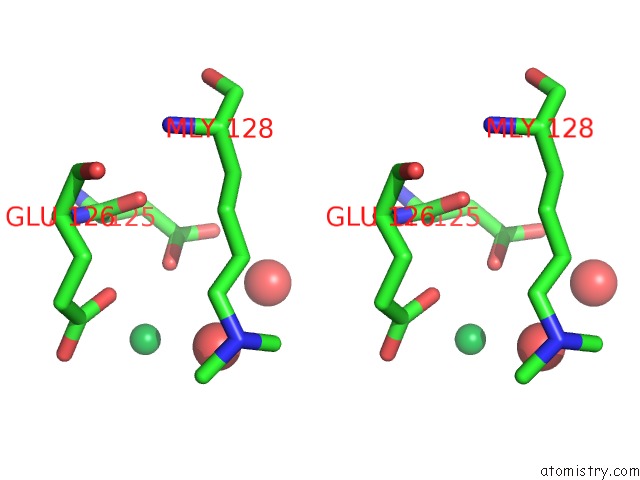
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 2 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 3 out of 9 in 1z1y
Go back to
Ytterbium binding site 3 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
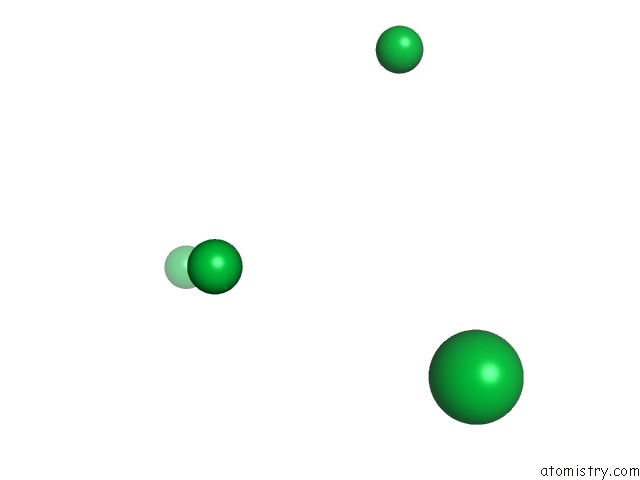
Mono view
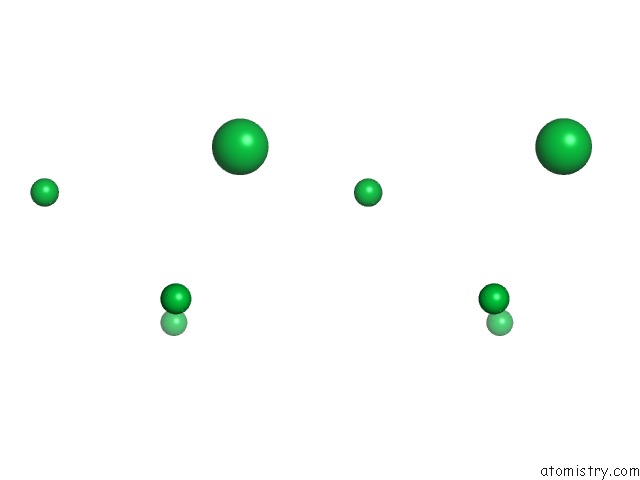
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 3 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 4 out of 9 in 1z1y
Go back to
Ytterbium binding site 4 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
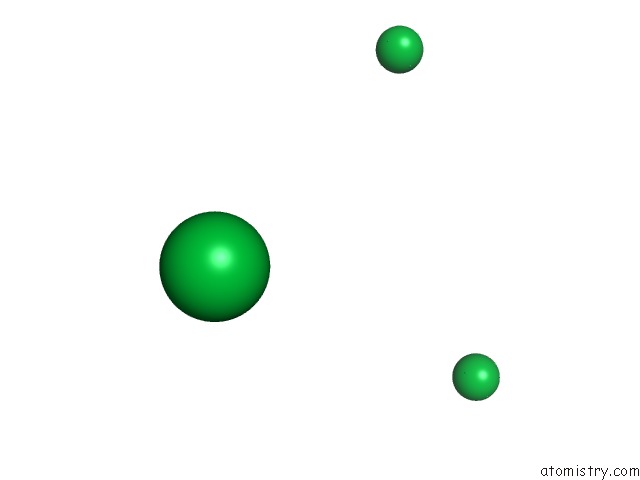
Mono view
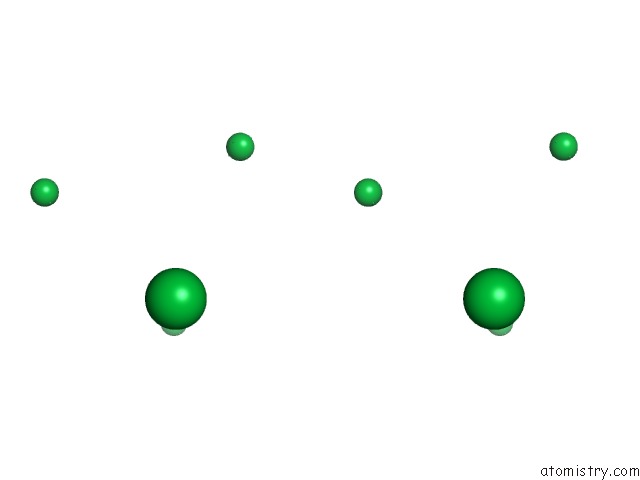
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 4 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 5 out of 9 in 1z1y
Go back to
Ytterbium binding site 5 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
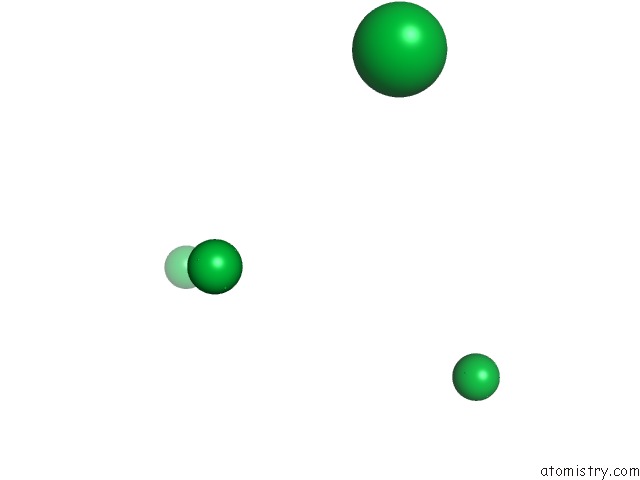
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 5 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 6 out of 9 in 1z1y
Go back to
Ytterbium binding site 6 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
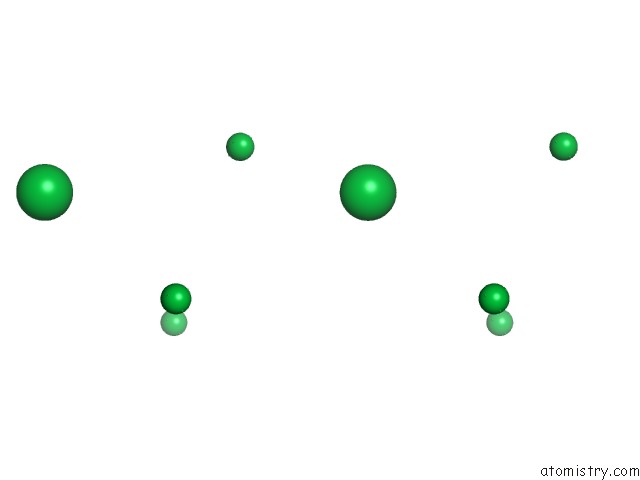
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 6 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 7 out of 9 in 1z1y
Go back to
Ytterbium binding site 7 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
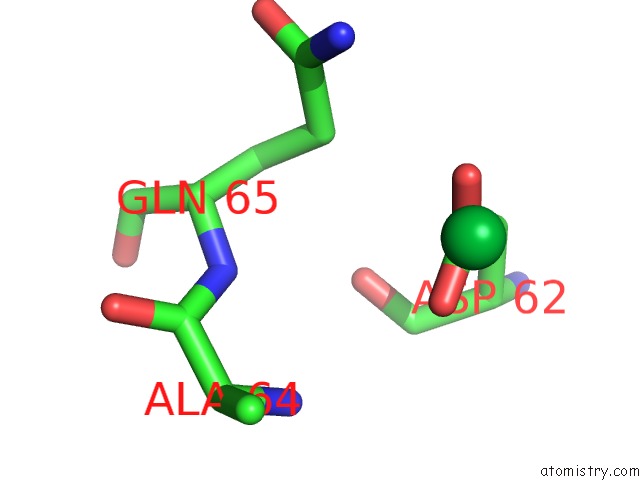
Mono view
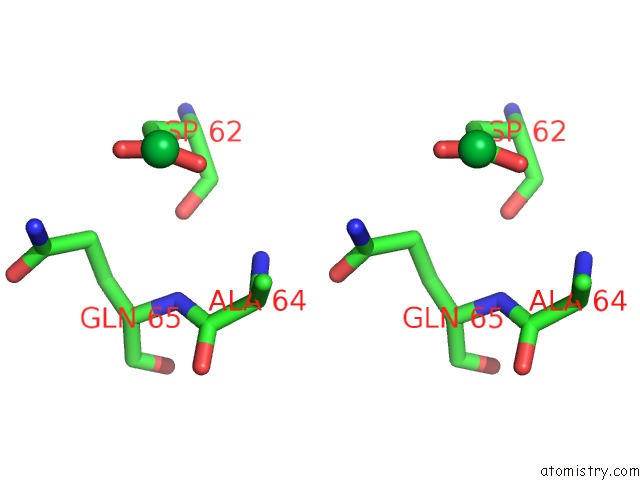
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 7 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 8 out of 9 in 1z1y
Go back to
Ytterbium binding site 8 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
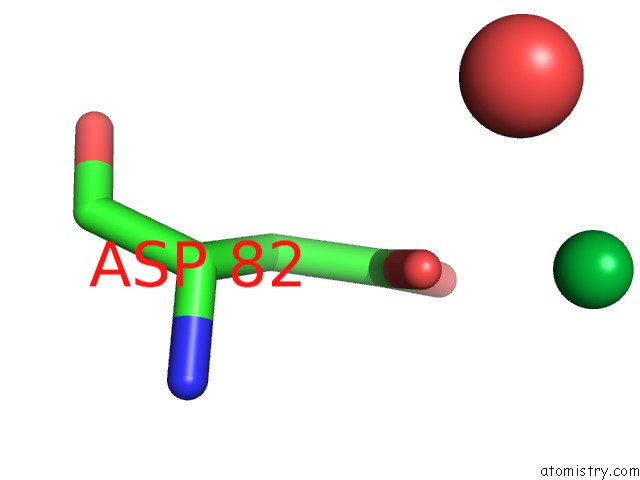
Mono view
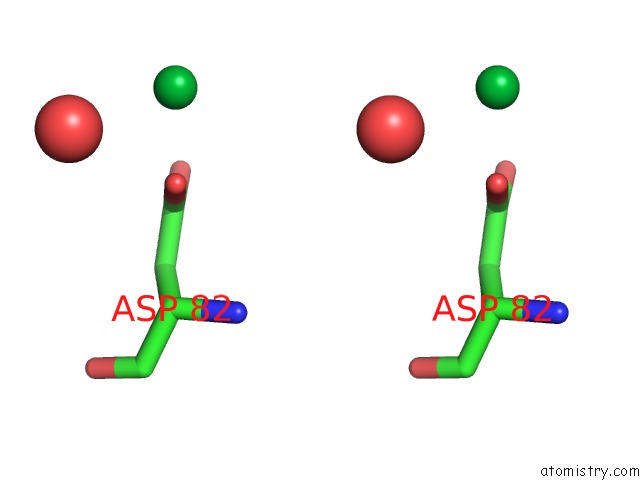
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 8 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Ytterbium binding site 9 out of 9 in 1z1y
Go back to
Ytterbium binding site 9 out
of 9 in the Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax
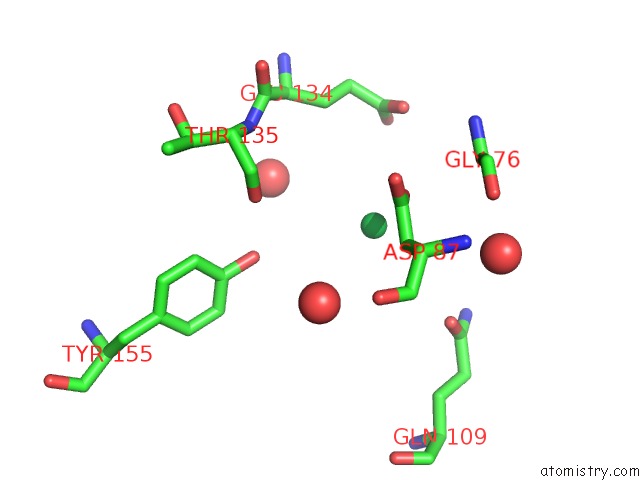
Mono view
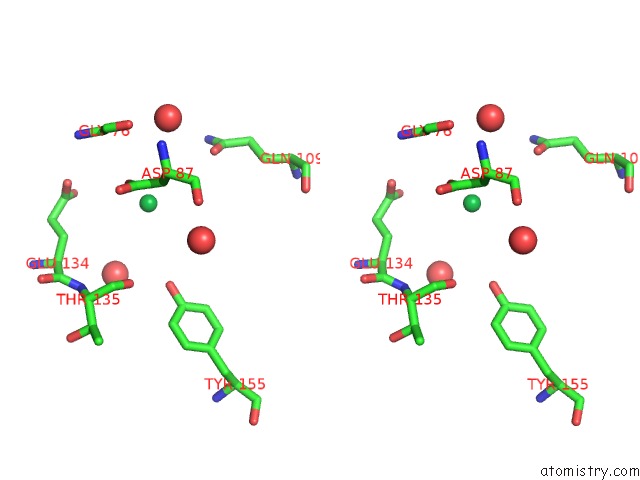
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 9 of Crystal Structure of Methylated PVS25, An Ookinete Protein From Plasmodium Vivax within 5.0Å range:
|
Reference:
A.K.Saxena,
K.Singh,
H.P.Su,
M.M.Klein,
A.W.Stowers,
A.J.Saul,
C.A.Long,
D.N.Garboczi.
The Essential Mosquito-Stage P25 and P28 Proteins From Plasmodium Form Tile-Like Triangular Prisms Nat.Struct.Mol.Biol. V. 13 90 2006.
ISSN: ISSN 1545-9993
PubMed: 16327807
DOI: 10.1038/NSMB1024
Page generated: Sat Oct 12 20:58:42 2024
ISSN: ISSN 1545-9993
PubMed: 16327807
DOI: 10.1038/NSMB1024
Last articles
Na in 1VQLNa in 1VQK
Na in 1VQ9
Na in 1VQ8
Na in 1VQ7
Na in 1VQ5
Na in 1VQ6
Na in 1VQ4
Na in 1VI6
Na in 1VKG