Ytterbium »
PDB 5l2m-8t0t »
8f7n »
Ytterbium in PDB 8f7n: Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
Protein crystallography data
The structure of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain, PDB code: 8f7n
was solved by
S.Salar,
F.D.Schubot,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.60 / 2.70 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 184.33, 184.33, 113.36, 90, 90, 90 |
| R / Rfree (%) | 24.4 / 26 |
Ytterbium Binding Sites:
The binding sites of Ytterbium atom in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
(pdb code 8f7n). This binding sites where shown within
5.0 Angstroms radius around Ytterbium atom.
In total 10 binding sites of Ytterbium where determined in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain, PDB code: 8f7n:
Jump to Ytterbium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Ytterbium where determined in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain, PDB code: 8f7n:
Jump to Ytterbium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Ytterbium binding site 1 out of 10 in 8f7n
Go back to
Ytterbium binding site 1 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
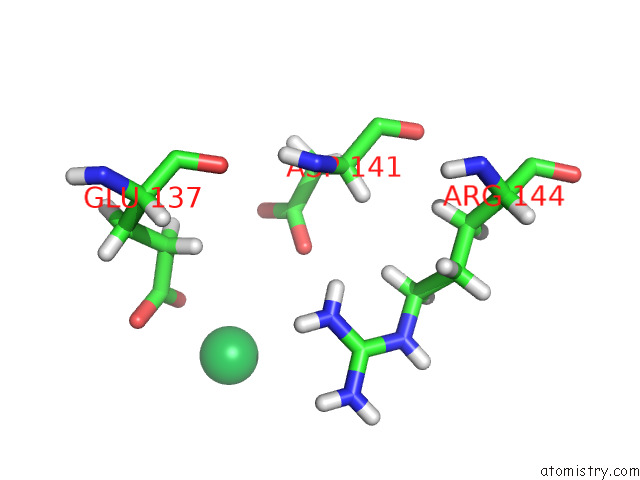
Mono view
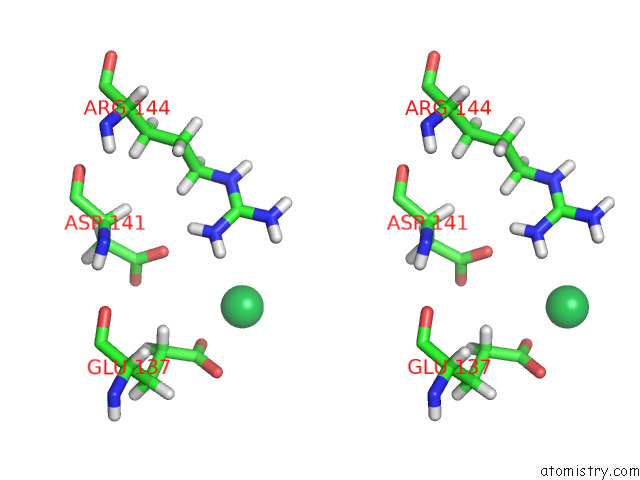
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 1 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 2 out of 10 in 8f7n
Go back to
Ytterbium binding site 2 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
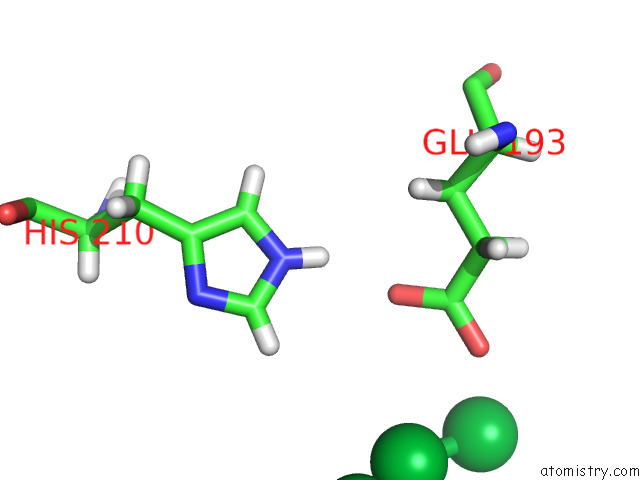
Mono view
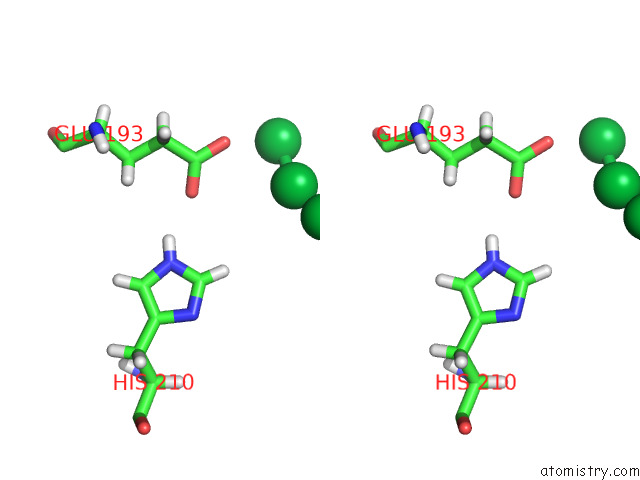
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 2 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 3 out of 10 in 8f7n
Go back to
Ytterbium binding site 3 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 3 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 4 out of 10 in 8f7n
Go back to
Ytterbium binding site 4 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 4 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 5 out of 10 in 8f7n
Go back to
Ytterbium binding site 5 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
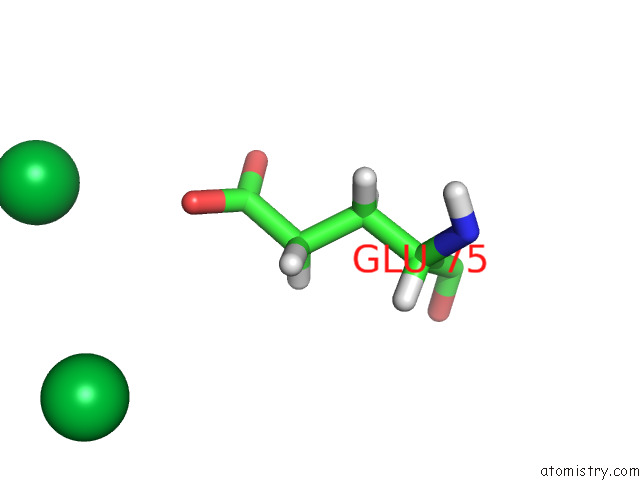
Mono view
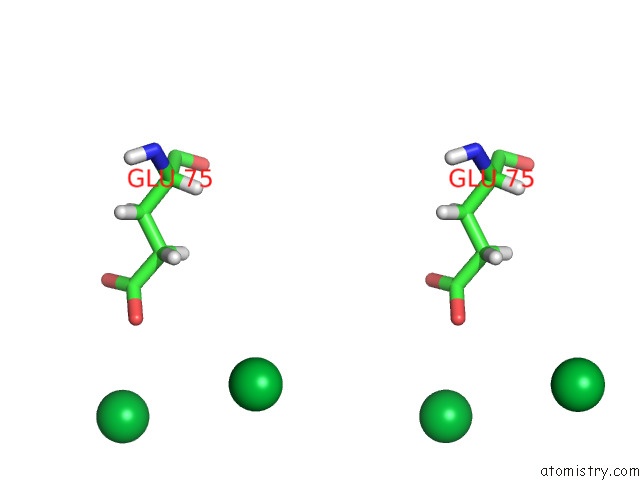
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 5 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 6 out of 10 in 8f7n
Go back to
Ytterbium binding site 6 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
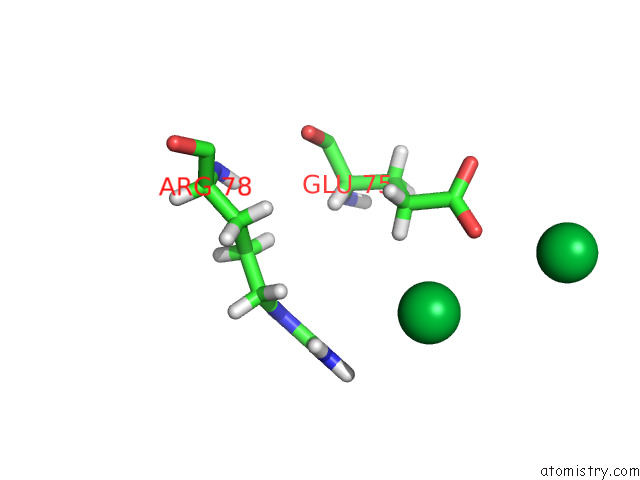
Mono view
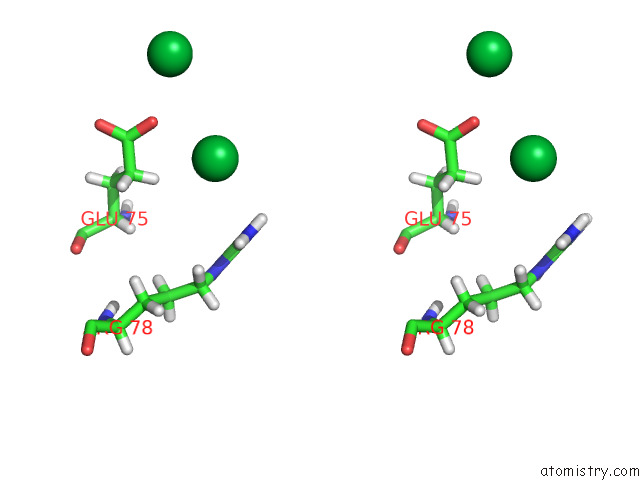
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 6 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 7 out of 10 in 8f7n
Go back to
Ytterbium binding site 7 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
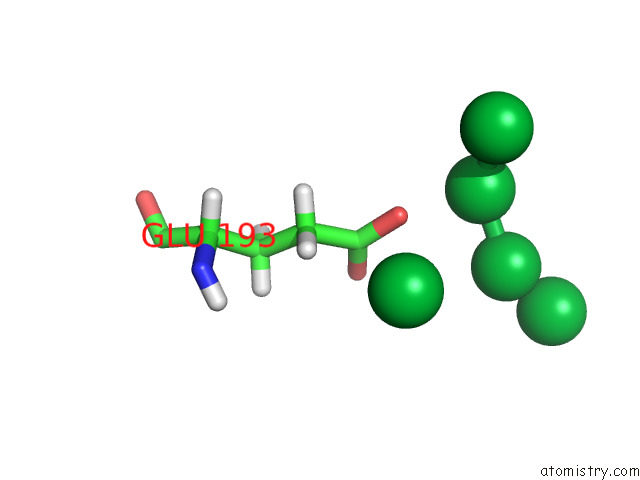
Mono view
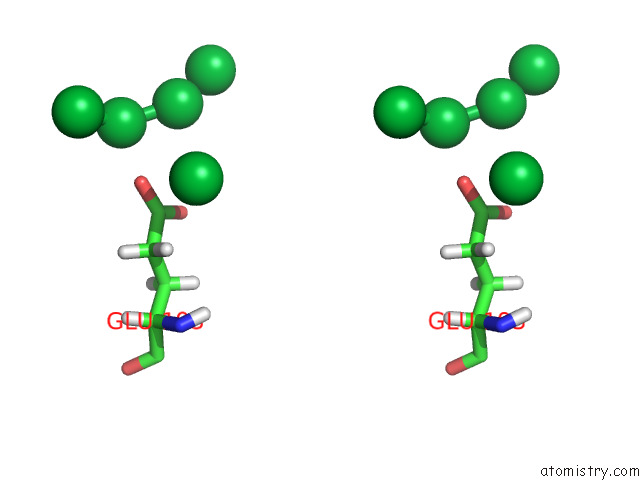
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 7 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Ytterbium binding site 8 out of 10 in 8f7n
Go back to
Ytterbium binding site 8 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
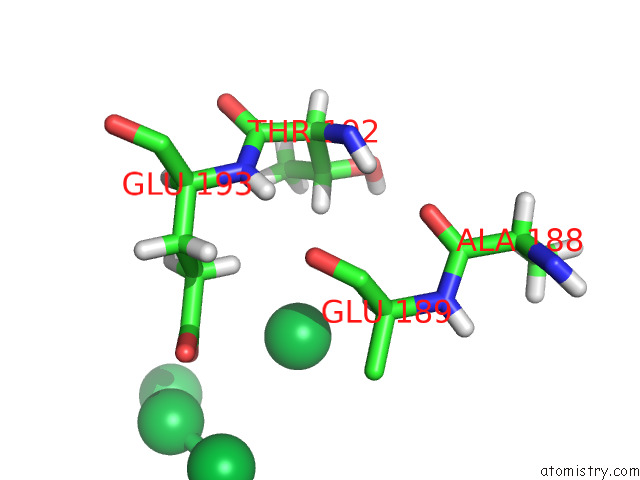
Mono view
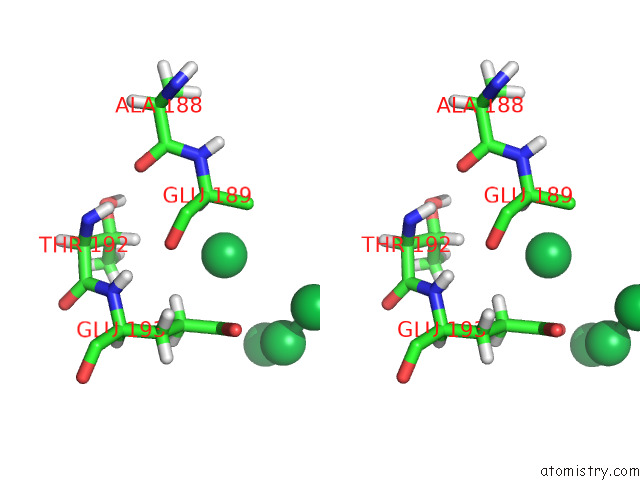
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 8 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
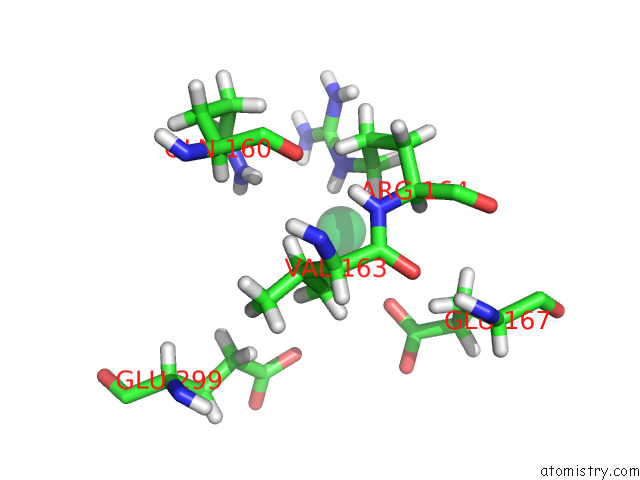
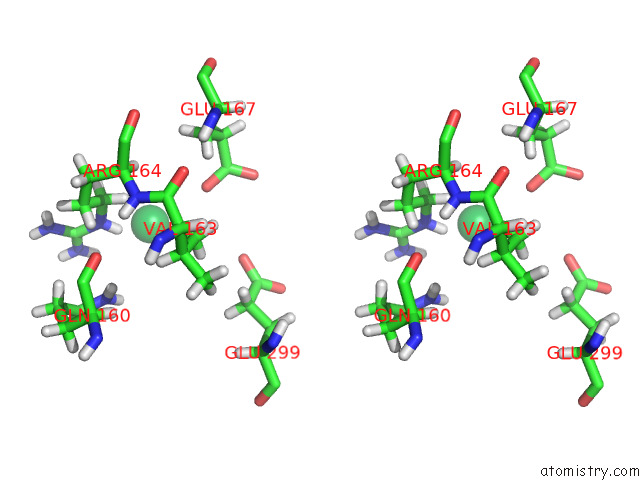
Ytterbium binding site 9 out of 10 in 8f7n
Go back to
Ytterbium binding site 9 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 9 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
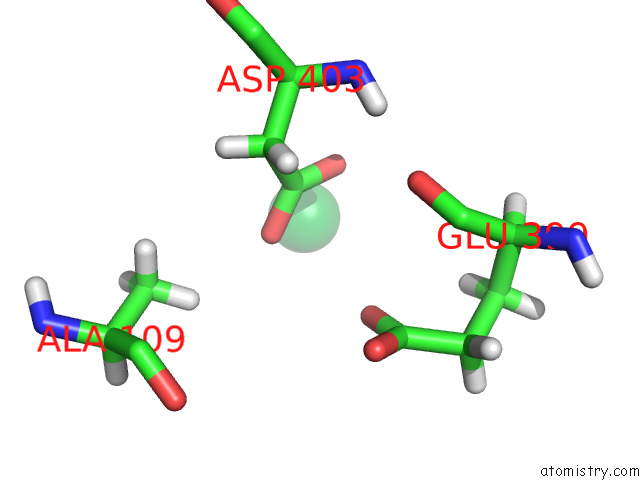
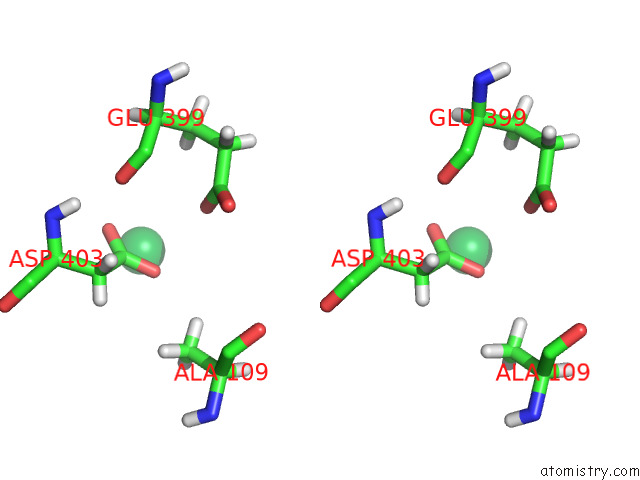
Ytterbium binding site 10 out of 10 in 8f7n
Go back to
Ytterbium binding site 10 out
of 10 in the Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Ytterbium with other atoms in the Yb binding
site number 10 of Crystal Structure of Chemoreceptor Mcpz Ligand Sensing Domain within 5.0Å range:
|
Reference:
S.Salar,
N.E.Ball,
H.Baaziz,
J.C.Nix,
R.C.Sobe,
K.K.Compton,
I.B.Zhulin,
A.M.Brown,
B.E.Scharf,
F.D.Schubot.
The Structural Analysis of the Periplasmic Domain of Sinorhizobium Meliloti Chemoreceptor Mcpz Reveals A Novel Fold and Suggests A Complex Mechanism of Transmembrane Signaling. Proteins 2023.
ISSN: ESSN 1097-0134
PubMed: 37213073
DOI: 10.1002/PROT.26510
Page generated: Sat Oct 12 21:29:29 2024
ISSN: ESSN 1097-0134
PubMed: 37213073
DOI: 10.1002/PROT.26510
Last articles
F in 7EPEF in 7EPF
F in 7EOU
F in 7EOR
F in 7EOB
F in 7ENN
F in 7EO9
F in 7EO4
F in 7EFN
F in 7EGP